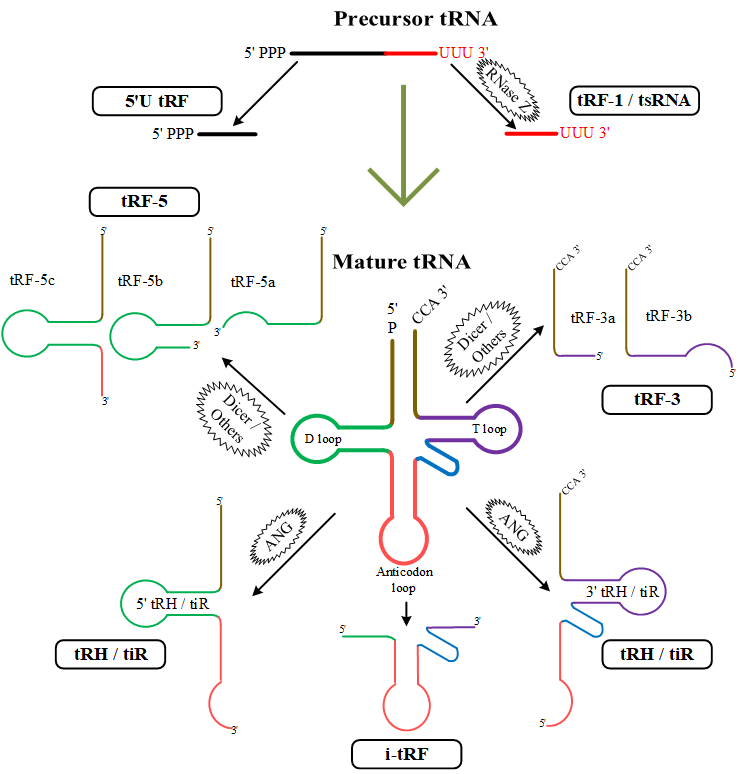
Transfer RNA-derived fragments (tRFs) are a new class of small noncoding RNA (ncRNA) in the length of 13-48 nucleotides. tRFs are derived from non-random cleavage of either the precursor or mature tRNA. According to the cleavage site of tRNA, tRFs are classified into 6 categories as shown in the figure below:
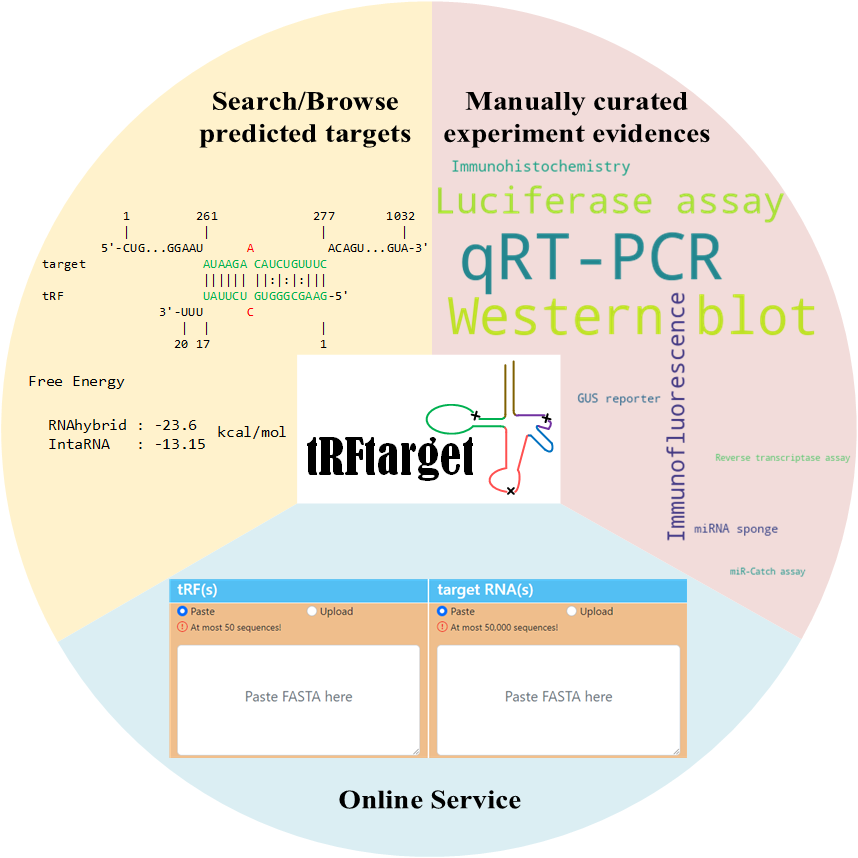
tRFtarget includes the computational predicted binding interactions between tRFs and target transcripts for multiple organisms based on the state-of-the-art prediction tools RNAhybrid and IntaRNA.
tRFtarget can provide detailed information about the binding interactions as well as an illustration of the interaction sequences. Furthermore, tRFtarget can be also searched by tRF ID, gene or transcript to get a comprehensive transcriptome-wide overview of tRF-target interactions.
tRFtarget also provides online service to allow users upload custom tRF and target sequences and get the predicted binding interactions using our pipeline tRFtarget-pipeline.
Besides, tRFtarget also integrates experimental evidence of the predicted tRF-RNA interactions based on manually curated publications. The experimental evidence are classed into 2 categories: gene-level evidence and site-level evidence.